Structure-function studies reveal ComEA contains an ... - Nature.com
Abstract
An essential step in bacterial transformation is the uptake of DNA into the periplasm, across the thick peptidoglycan cell wall of Gram-positive bacteria, or the outer membrane and thin peptidoglycan layer of Gram-negative bacteria. ComEA, a DNA-binding protein widely conserved in transformable bacteria, is required for this uptake step. Here we determine X-ray crystal structures of ComEA from two Gram-positive species, Bacillus subtilis and Geobacillus stearothermophilus, identifying a domain that is absent in Gram-negative bacteria. X-ray crystallographic, genetic, and analytical ultracentrifugation (AUC) analyses reveal that this domain drives ComEA oligomerization, which we show is required for transformation. We use multi-wavelength AUC (MW-AUC) to characterize the interaction between DNA and the ComEA DNA-binding domain. Finally, we present a model for the interaction of the ComEA DNA-binding domain with DNA, suggesting that ComEA oligomerization may provide a pulling force that drives DNA uptake across the thick cell walls of Gram-positive bacteria.
Introduction
Natural competence for transformation is a mechanism of horizontal gene transfer widespread in both Gram-positive and Gram-negative bacteria, as well as in some archaea (for reviews see1,2). The transformation process consists of two major steps: uptake and transport (Fig. 1A). Uptake is the movement of environmental transforming DNA (tDNA) into the periplasm, across the thick peptidoglycan cell wall of Gram-positive bacteria or the outer membrane and thin peptidoglycan layer of Gram-negative bacteria. Transport is the subsequent translocation of single-stranded tDNA from the periplasm to the cytoplasm where it can recombine with the chromosome to generate a transformant. The structure-function studies presented here focus on the first step, DNA uptake.
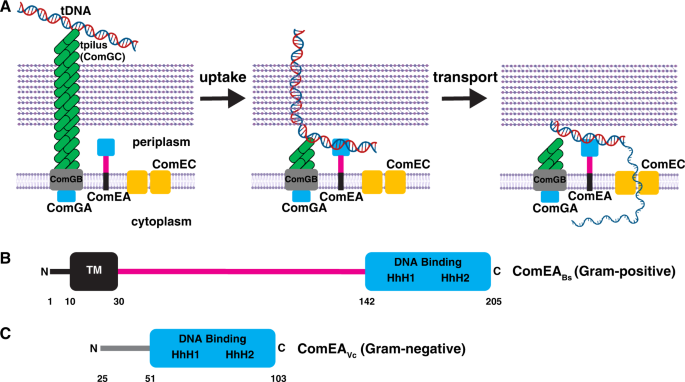
A The tpilus is composed of ComGC, whose assembly requires the ATPase ComGA. The tpilus is believed to be anchored to the membrane protein ComGB. The tpilus binds weakly to DNA and retracts to pull it into the periplasm. Here, the DNA encounters ComEA, which stabilizes binding to the cell and propels continued uptake of the DNA. ComEC is proposed to degrade one strand of DNA and provide the channel for transport of DNA into the cytoplasm. B Domain architecture of ComEA from a representative Gram-positive bacterium, B. subtilis. C Domain architecture of ComEA from a representative Gram-negative bacterium, V. cholerae. Residues 1–24 are not shown in order to highlight the fact that they comprise a predicted secretion signal that is cleaved to generate mature ComEAVc, which diffuses freely in the periplasm. TM, predicted transmembrane region. The magenta line denotes a region of unknown function, which is addressed in this study. HhH, helix-hairpin-helix motifs. Residue numbering corresponds to ComEABs or ComEAVc. Elements of the figure were created with BioRender.com.
Uptake is initiated at the cell surface when tDNA interacts with a transformation pilus (tpilus). Following this contact with the tpilus, the movement of tDNA into the periplasm is driven by its interaction with the widely conserved periplasmic protein ComEA (Fig. 1A). ComEA, was discovered in genetic screens for transformation deficiency in Bacillus subtilis. It was subsequently shown to be a non-specific DNA-binding protein required for both stable binding of tDNA to the cell and for tDNA uptake3,4. In B. subtilis and other Gram-positive bacteria, ComEA is bound to the membrane by an N-terminal transmembrane region4. This membrane anchor and a C-terminal DNA binding domain are separated by a region of unknown function consisting of ~110 amino acids (Fig. 1B). In sharp contrast, ComEA in the Gram-negatives contains only a single (stand-alone) DNA-binding domain which is free to diffuse in the periplasm5,6,7 (Fig. 1C).
In both Gram-negative and positive bacteria, except for Helicobacter pylori8, uptake is initiated by type 4 pili that bind DNA and retract, pulling a segment of transforming DNA into the periplasmic compartment9,10. It has been proposed that in the Gram-negative Vibrio cholerae and Neisseria gonorrhoeae, the single-domain ComEA then binds to this introduced tDNA segment, preventing loss of tDNA by backward diffusion across the outer membrane6,11. Thus, the pilus initiates uptake while ComEA operates as a Brownian ratchet12, providing the driving force for uptake of the bulk of the tDNA into the periplasm. In all bacteria, following uptake to the periplasm, one strand of tDNA is degraded and the remaining strand is transported to the cytoplasm through the ComEC membrane channel (reviewed in1,2). In Gram-positives, the membrane-associated ATPase ComFA and its partner protein ComFC, appear to provide the energy for transport. Equivalent Gram-negative proteins have not been identified.
The complex structure and membrane anchoring of ComEA in the Gram-positives suggests that it may act differently than the simple Brownian ratchet mechanism proposed for V. cholerae and N. gonorrhoeae. To investigate this complex structure and to gain insight into its mode of action, we conducted an in vitro and in vivo structure-function investigation, centered on the B. subtilis protein. Here we present X-ray crystal structures of ComEA from B. subtilis (ComEABs) and Geobacillus stearothermophilus (ComEAGs) together with complementary genetic and biophysical analyses. These studies reveal that the DNA binding domain is an atypical helix-hairpin-helix domain, and, most importantly, that a previously unidentified domain lies within the Gram-positive ComEA region of unknown function. We show that this domain drives ComEA oligomerization, and that oligomerization is required for genetic transformation. We postulate that the unexpected role of ComEA oligomerization in DNA uptake is explained by DNA-protein condensation providing a driving force for tDNA uptake across the thick cell wall of Gram-positive bacteria.
Results
X-ray crystal structures of ComEABs and ComEAGs
To gain mechanistic insight into the function of ComEA, we cloned, overexpressed, purified, and determined the X-ray crystal structures of ComEABs and ComEAGs to 3.20 Å and 3.05 Å resolution, respectively (Figs. 2A, 2B, and Table S1). No electron density was evident for the ComEABs DNA-binding domain or linker region, but there was clear density corresponding to a previously undescribed domain (amino acids 60–122) (Fig. 2A). There are seven of these domains arranged head-to-tail in the ComEABs crystallographic asymmetric unit, and we named this region the oligomerization domain (OD) (Fig. 2A).
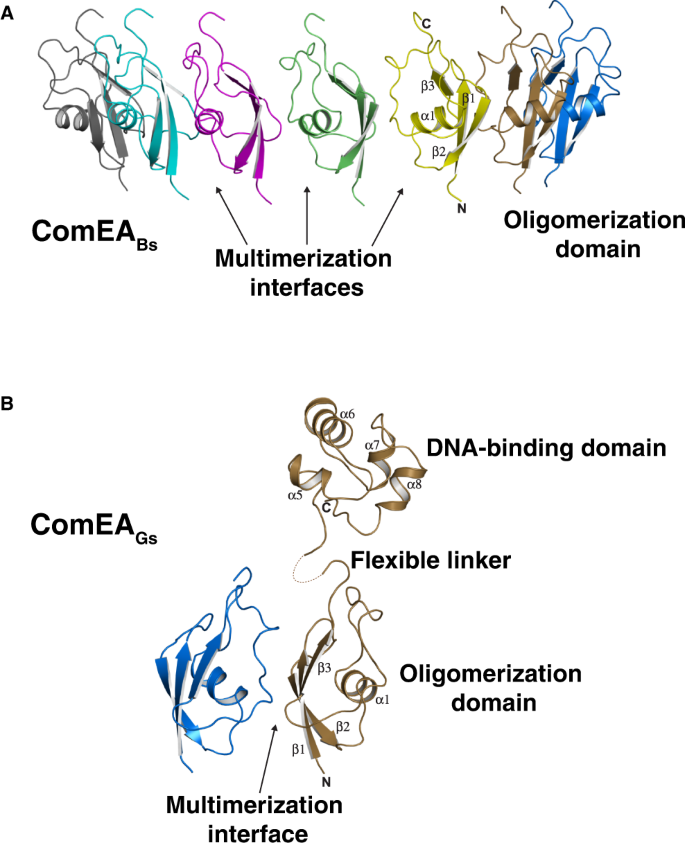
A) X-ray crystal structure of ComEABs. One asymmetric unit containing seven protomers is shown. Arrows here point to only three of the six multimerization interfaces in the asymmetric unit. B) X-ray crystal structure of ComEAGs. One asymmetric unit containing two protomers is shown.
In contrast to ComEABs, in the ComEAGs structure there was interpretable electron density corresponding not only to the OD (amino acids 62–124) but also to one DNA-binding domain (amino acids 143–207) per asymmetric unit, which contained two OD protomers again arranged head-to-tail (Fig. 2B). Because there is no interpretable electron density corresponding to the ComEAGs linker region (amino acids 125–142) of either protomer, it is unknown which of the OD monomers in the asymmetric unit OD dimer connects to the single modeled DNA-binding domain. In fact, we could model the DNA-binding domain in the ComEAGs structure only because it made fortuitous crystal contacts. We conclude from the ComEA structures that the linker domain is flexible, that there are minimal contacts between the OD and DNA-binding domains, and that the ODs have the potential to form multimers in solution.
We note that OD multimerization extends beyond the asymmetric unit (Fig. 2A, B). More specifically, in the ComEABs structure, the seven-membered asymmetric unit uses the OD multimerization interface to form a 14-membered head-to-tail ring around the crystallographic two-fold axis (Fig. 3A, B). Similarly, in the ComEAGs structure, examination of symmetry mates reveals that the two-membered asymmetric unit uses the OD multimerization interface to form a kinked-ring spiral along the crystallographic screw axis, containing 12 head-to-tail protomers per turn (Fig. 3C, D). A slight tilt between ComEAGs protomers (Fig. S1) has minimal effects on the ComEA inter-dimer bonds, but it causes ComEAGs to form a kinked-ring rather than a closed ring within the crystals.
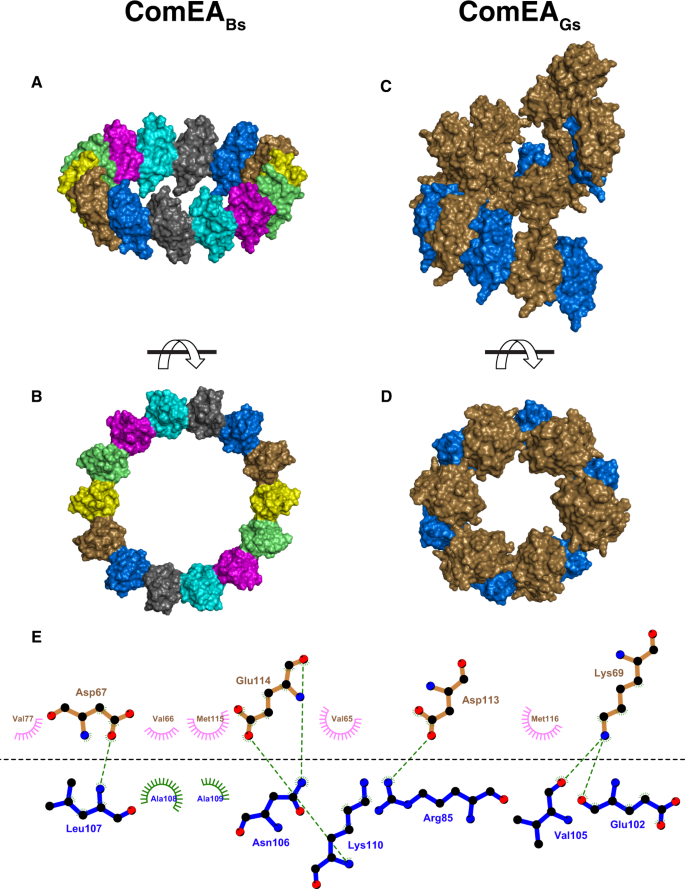
A and B Front and side views of the ComEABs crystallographic ring, respectively. C and D Front and side views of the ComEAGs crystallographic kinked ring, respectively. E Schematic representation of the ComEAGs multimerization interface. ComEA chains A and B are depicted as blue and brown bonds, respectively. Hydrogen bonds are depicted as green dashed lines. Hydrophobic contacts are depicted as lines radiating from the semicircles and spheres. The schematic was produced with LigPlot+50.
ComEA oligomerization interface amino acids are conserved (Fig. S2) and mediate conspicuous intermolecular interactions between the OD domains of ComEA protomers, e.g., the side chains of ComEAGs Arg85 and Asp113 (corresponding to ComEABs Arg83 and Asp111) which form intermolecular salt bridges, (Fig. 3E). This suggested that ComEA may oligomerize not only in the crystals but also in solution and we proceeded to explore this possibility.
ComEA oligomerizes in solution
To confirm that the OD observed in both ComEA X-ray crystal structures mediated reversible self-association in solution, we analyzed different concentrations of ComEAGs using sedimentation velocity analytical ultracentrifugation (SV-AUC)13. At 10.3 μM, the majority of ComEAGs has a sedimentation coefficient of 1.6 S, consistent with the ComEA monomer. However, at 157 μM, the majority of ComEAGs has a sedimentation coefficient of 2.2 S, suggesting reversible dimerization as a function of mass action (Fig. 4A). At an intermediate concentration of 31.3 μM we observed both monomer and dimer, and were able to fit the sedimentation velocity data to a discrete monomer-dimer equilibrium model13, resulting in a Kd of 33.8 μM (95% confidence intervals: 19.3 μM, 48.4 μM) (Table S4). The apparent ComEA dimerization affinity is likely an underestimation of its in vivo dimerization affinity, because diffusion of ComEA in vivo is limited to two dimensions in the cell membrane.
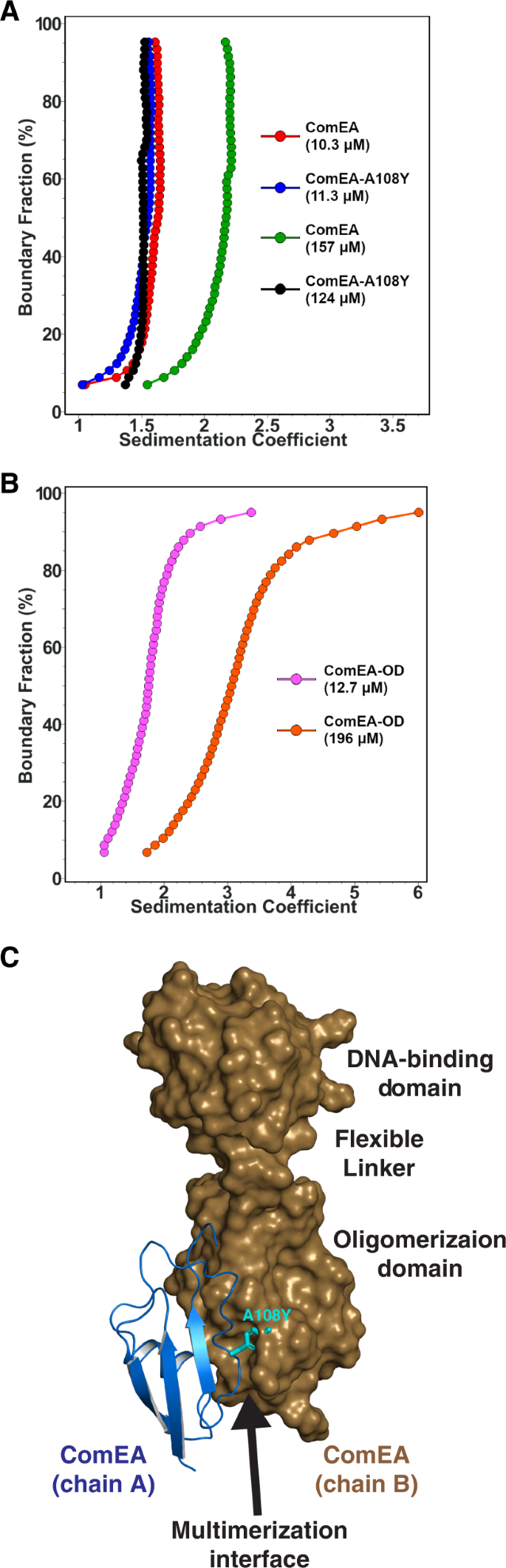
A Integral sedimentation coefficient distribution overlays comparing the dimerization potential of ComEAGs at 10.3 μM (red) and 157 μM (green) and ComEAGs-A108Y at 11.3 μM (blue) and 124 μM (black). Only ComEAGs dimerizes at higher concentration, while ComEAGs-A108Y remains monomeric. B Integral sedimentation coefficient distribution overlays of the ComEAGs OD at 12.7 μM (magenta) and 196 μM (orange), showing reversible self-association. C Structure of ComEAGs with chain A depicted as a cartoon and chain B depicted as a surface. ComEAGs chain A Ala108 was mutated to Tyr and is depicted as magenta sticks.
Consistent with the SV-AUC analysis of ComEAGs, a truncated ComEAGs protein consisting of the OD alone (ComEAGs-OD) also multimerized in solution (Fig. 4B). However, ComEAGs-OD formed primarily monomers and dimers at low concentration (12.7 μM) and larger oligomers with a maximum molar mass around 100 kDa at high concentration (196 μM) (Fig. S3). One possible kinetic explanation for the different multimeric states of ComEAGs and ComEAGs-OD at similar concentrations is that the presence of the ComEAGs DNA-binding domain slows the OD multimerization search, limiting collisions, thus slowing the OD multimerization rate (kon).
To confirm that ComEA multimerization is driven by the OD, we examined the X-ray crystal structures to identify a small amino acid buried in the OD multimerization interface that we could replace with a larger amino acid, introducing steric bulk and disrupting multimerization. We identified ComEAGs Ala108 (corresponding to ComEABs Ala106) and substituted it with tyrosine (Figs. 3E, 4A, C, and S4). Indeed, ComEAGs-A108Y was monomeric at both 11.3 μM and 124 μM (Fig. 4A). These SV-AUC experiments demonstrate that the OD drives ComEA multimerization in solution as predicted from the X-ray crystal structures.
ComEA oligomerizes on DNA
We further speculated that ComEA multimerization plays an important role in its interaction with DNA. To test this, we used multi-wavelength analytical ultracentrifugation (MW-AUC)14,15,16,17 to measure the binding of wild-type ComEAGs and ComEAGs-A108Y to DNA. MW-AUC is a novel technique that permits the spectral separation of protein and DNA species based on their unique absorbance spectra. Consequently, it is possible to measure the molar stoichiometry of the complexes formed and identify the type of macromolecule(s) forming each hydrodynamic species. The hydrodynamic measurements depend on each species' molar mass and hydrodynamic radius.
To elucidate the interactions between ComEA and double-stranded DNA, we mixed wild-type and mutant ComEA with a 14-bp double-stranded DNA duplex at a 5:1 and a 10:1 molar protein excess and a 10:1 protein excess over a 40-bp double-stranded DNA duplex (Fig. 5A–C and Table S2). In all cases, the DNA concentration was held constant at 1.5 μM, and a well-defined complex was formed, with less than 10% of the DNA remaining free in the solution. The deconvoluted DNA sedimentation pattern suggests the presence of a saturated, homogeneous complex being formed in all cases. As expected, the DNA sequestered excess protein until it was fully occupied, and the 14-bp sequence accommodated fewer ComEA molecules than the 40-bp sequence.
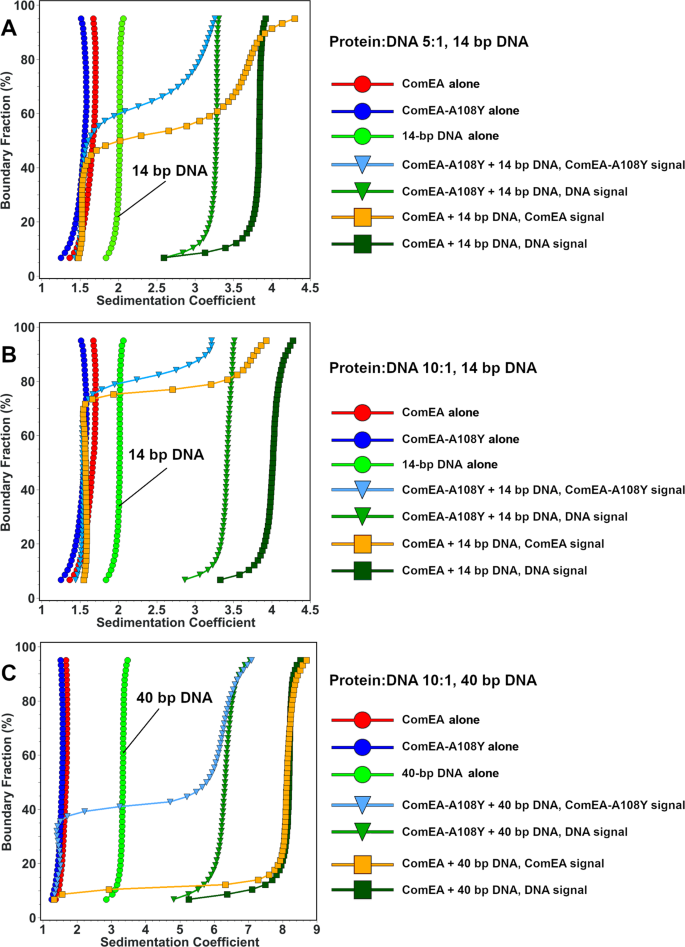
A and B Mixtures 5:1 and 10:1 molar ratios of wild-type ComEAGs and ComEAGs-A108Y with the 14-bp DNA molecule, respectively. Here, the DNA signals still suggest full saturation with protein, but the protein signals show more heterogeneous sedimentation coefficient distributions, consistent with more rapid exchange with the protein-DNA complex, which suggests a faster koff rate. More than 90% of the DNA is complexed with protein, shifting the 14 bp DNA distribution from 2.0 S for the control by itself to 3.3 S for the mutant, and 3.9 S for the wild-type in panel A. In panel B, increasing the protein concentration to 10:1, marginally shifts the DNA sedimentation for the wild-type further to about 4.0 S, while barely affecting the DNA sedimentation when mixed with the mutant. C Integral sedimentation coefficient distribution overlays for the deconvoluted protein and DNA signals from the 10:1 mixture of ComEAGs and ComEAGs-A108Y with 1.5 μM of the 40-bp DNA molecule. Unbound ComEA in the presence of DNA co-sediments with ComA in the absence of DNA. Again, more than 90% of the DNA signal shifts from the position of free 40 bp DNA at 3.3 s to a homogeneous composition at 6.3 S for the mutant, and 8.1 S for the wild-type, suggesting saturation of the DNA with ComEA. The ComEA signal closely tracks the DNA signal, suggesting a tight complex formation with a slow koff rate. For all plots, reference controls of each molecule by itself are shown as circles (ComEA: red circles, ComEA-A108Y: blue circles, 14 or 40 bp DNA, as indicated: green circles), symbols for interactions between DNA and wildtype protein are shown as squares (ComEA signal: orange squares, DNA signal: dark green squares) and interactions between ComEA-A108Y and DNA are shown in triangles (ComEA-A108Y signal: olive triangles, DNA signal: light blue triangles).
The sedimentation coefficient of the complex differed for wild-type and mutant and depended on the length of the DNA. The 14-bp double-stranded DNA duplex sedimented at 2.0 S in isolation but formed a 3.3 S complex with ComEAGs-A108Y, and a 3.9 S complex with wild-type ComEAGs in the 5:1 mixture (Fig. 5A). For this mixture, we measured a ~3:1 protein:DNA ratio for the complex formed with ComEAGs, and a ~2:1 ratio for the complex formed with the ComEAGs-A108Y protein. For the 10:1 protein:DNA mixture, a slightly larger 3.5 S complex was formed with ComEAGs-A108Y, a 4.0 S complex was formed with wild-type ComEAGs, and the protein:DNA ratios did not change significantly (Fig. 5B).
The 40-bp DNA sediments at 3.3 S when examined by itself, but, when mixed in a 10:1 ratio with ComEAGs-A108Y resulted in a 6.3 S complex, while the same mixture with wild-type ComEAGs resulted in a 8.1 S complex (Fig. 5C). For the 40-bp DNA, the difference between protein:DNA molar ratios of the formed complexes was more pronounced between wild-type and mutant. The wild-type ComEAGs displayed an approximately 7:1 ratio, but the complex between DNA and ComEAGs-A108Y suggested the presence of about a 4.7:1 ratio. While the accuracy of these ratios depends on the estimated molar extinction coefficients in the formed complexes, there is a clear difference in the amount of ComEA bound to DNA for the wild-type and mutant proteins.
In sum, these results suggest that wild-type ComEA, capable of oligomerizing, forms larger complexes with DNA than the A108Y mutant. Oligomerization facilitates efficient and cooperative packing of ComEA on DNA, which is further supported by gel shift analysis (detailed below).
ComEA oligomerization is required for transformation
The conservation in Gram-positive bacteria of both the ComEA OD domain itself and, significantly, the residues buried in the multimerization interface (Figs. S2 and 3E), led us to hypothesize that the OD is important for transformation. We measured this in vivo by comparing B. subtilis transformation in strains containing wild-type ComEABs; ComEABs-D111N, which disrupts a conserved intermolecular salt-bridge between D111 and R83 (corresponding to ComEAGs Asp113 and Arg85, Fig. 3E); ComEABs-A106Y (corresponding to ComEAGs-A108Y), which, as shown above, sterically blocks OD multimerization (Fig. 4A, C); and ComEA-ΔOD, containing a deletion of the entire OD, while leaving intact both the transmembrane region and the DNA-binding domain. In comparison to wild-type ComEABs, ComEABs-D111N reduced B. subtilis transformation nearly 100-fold (Fig. 6A). Even more striking were the ComEABs-A106Y and ComEA-ΔOD mutations that reduced B. subtilis transformation about 1,000-fold, a phenotype equivalent to that of a complete comEA deletion. Western blot analysis showed that the wild-type and mutant ComEA proteins were similarly expressed in B. subtilis (Fig. 6B). To determine whether the A106Y oligomerization deficient mutant exerts its effect on transformation by preventing DNA uptake to the periplasm, we used rhodamine-labeled tDNA, which can be taken up into the periplasm but cannot cross the cell membrane18. As shown in (Fig. 6C), the A106Y mutation prevents stable DNA association with competence-expressing cells, which we have shown previously required uptake to the periplasm18. We conclude that ComEA oligomerization plays an unexpected and necessary role in the transformation of B. subtilis and likely of other Gram-positive bacteria as well.
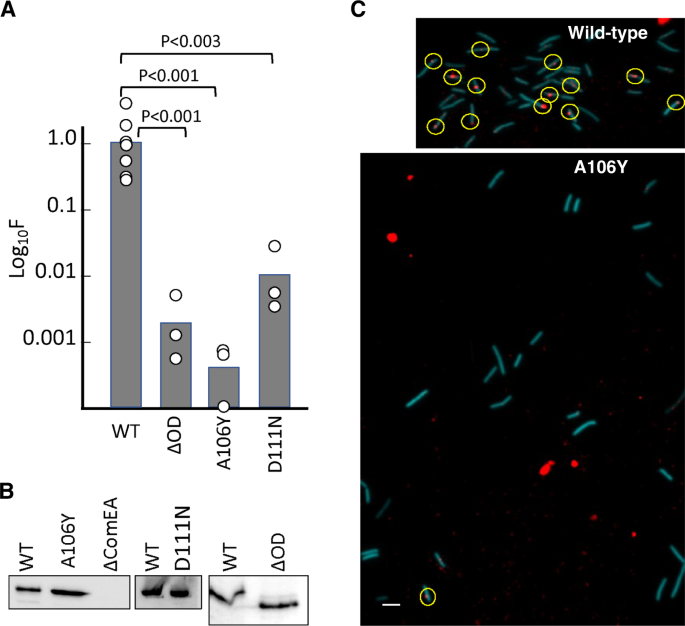
All mutations were introduced into the chromosome of BD9007, which carries a competence-specific fusion of a sequence encoding cyan fluorescent protein (CFP) expressed from the promoter of comG, placed at the ectopic amyE locus to enable the identification of competence-expressing cells by epifluorescence microscopy. Transformation experiments were performed as biological triplicates and average frequencies, normalized to the wild-type values are plotted as bar graphs in panel A with the data points included. The larger number of data points for the wild-type control reflects the inclusion of this strain in all three biological replicates. The mean normalized values with standard deviations for the mutants were 0.0012 ± 0.00047 (ΔOD), 0.0.0005 ± 0.00024 (A106Y), 0.11 ± 0.03 (D111N). The p values were determined using two-sided t-tests. Panel B shows Western blots for the corresponding wild-type and mutant extracts using anti-GFP antiserum. ΔcomEA extracts were included as controls for the identity of the ComEA signals. Panel C shows typical epifluorescent images from the wild-type (BD9007) and its isogenic A106Y mutant equivalent, after transformation with rhodamine-labeled lambda bacteriophage DNA for 45-min. Competence-expressing cells were identified by CFP fluorescence (pseudocolored cyan) and detectable cell-associated rhodamine-tDNA signals are circled. For the wild type, 13 out of 42 cells exhibited at least one red dot, while for the 23 mutant cells only one barely detectable cell-associated dot was observed. The large red blotches in the lower panel are not associated with cells and are due to contaminating fluorescent material of unknown origin. The scale bar on the lower left of the image corresponds to 1 micron. The microscopy and Western blotting experiments in panels B and C were each repeated three times with closely similar results. Source data are provided as a Source Data file.
ComEA-DNA interactions are required for transformation
The ComEAGs X-ray crystal structure revealed not only the structure of the OD, but the three-dimensional shape of the ComEA DNA-binding domain. As predicted from sequence analysis and noted previously6, the ComEA DNA-binding domain structure contains two helix-hairpin-helix (HhH) motifs (Figs. 1B and 7A). Structures of canonical HhH-containing proteins showed that pairs of HhH motifs are typically connected by an α-helix, and these five helices together are referred to as (HhH)2 domains19. This linker helix that connects the HhH motifs is conspicuously absent in ComEA, and its two HhH motifs are instead connected by a loop (Fig. 7B). The ComEA DNA-binding domain is, therefore, an atypical HhH domain that forms a well-folded and compact DNA-binding core without the use of a connector helix.
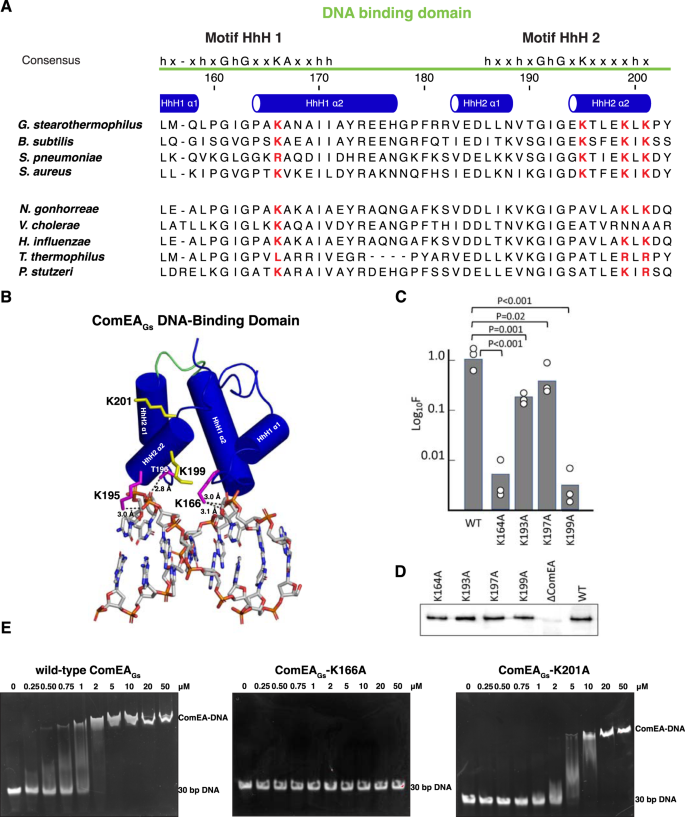
A Alignments of the HhH motifs from ComEA proteins expressed by four Gram-positive (top) and Gram-negative (bottom) bacteria that have been used for transformation studies. The four lysine residues chosen for mutagenesis are highlighted in red. Secondary structure assignments were derived from the crystal structure of ComEAGs. B Model of the ComEAGs DNA-binding domain (α-helices depicted as cylinders) in complex with dsDNA (depicted as sticks). The loop connecting the HhH1 and HhH2 is colored green. Hydrogen bonds are depicted as black dashed lines. For clarity, ComEAGs is depicted in complex with 8 bp central to the complex rather than the 15 bp used in refinement. C Transformation frequencies for the HhH motif mutants. Transformation experiments were performed as biological triplicates and average frequencies, normalized to the wild-type (BD9007) values are plotted as bar graphs with the data points included. The mean normalized values with standard deviations for the mutants were 0.0054 ± 0.004 (K164A), 0.23 ± 0.066 (K193A), 0.54 ± 0.22 (K197A), 0.0031 ± 0.0025 (K199A). The p values were determined using two-sided t-tests. D shows Western blots for the wild-type and mutant ComEA proteins obtained using anti-GFP antibody. This Western blot experiment was repeated a total of three times with nearly identical results. E EMSA analysis of wild-type ComEAGs, ComEA-K166A, and ComEA-K201A. The EMSA analysis was repeated at least two times with nearly identical results. Source data are provided as a Source Data file.
To explore ComEA-DNA interaction, we used Dali20 and HADDOCK 2.421 to generate a ComEAGs-DNA model (Fig. 7B). In addition to numerous hydrogen bonds between protein backbone nitrogens and DNA phosphate groups, which is typically how HhH domains interact with DNA19,22, we observed that the sidechains of Lys166, Lys195, and Thr196 formed hydrogen bonds with DNA phosphate groups. To begin to understand the importance of ComEA sidechain interactions to DNA binding and ComEA function, we mutated some of the corresponding interfacial residues in ComEABs (e.g., ComEABs K164 and K193) to alanine and tested their effect on transformation in B. subtilis (Fig. 7C). Furthermore, in the absence of an experimentally-determined ComEA-DNA structure, we considered the possibility that the in silico model was incomplete. For example, the ComEA-DNA model does not show possible interactions between ComEA DNA-binding domains that might occur when oligomers of ComEA bind DNA. Thus, we mutated additional ComEABs residues K197 and K199 (equivalent to ComEAGs residues K199 and K201) to alanine and tested their effects on transformation in B. subtilis.
ComEABs mutants K164A, K193A, K197A, and K199A were individually expressed from the chromosome in B. subtilis and their effects on transformation were compared to that of wild-type ComEA (Fig. 7C). Western blot analysis showed that wild-type ComEABs and the ComEABs proteins containing mutations in the HhH motifs were expressed to similar levels (Fig. 7D). These in vivo studies also showed that the K164A and K199A mutations caused more than a 2-log decrease in transformation (Fig. 7C), in contrast to the reproducible 2-4-fold effects of K193A and K197A.
In the ComEAGs-DNA model, residue K166 (equivalent to ComEABs K164) contacts DNA. Consistent with this observation and the ComEABs-K164 loss-of-function for transformation in vivo (Fig. 7C), purified ComEAGs-K166A (Fig. S4) does not bind DNA as determined using an electrophoretic mobility shift assay (EMSA) (Fig. 7E). In contrast, in the ComEAGs-DNA model, residue K201 (equivalent to ComEABs K199) does not contact DNA (Fig. 7B). ComEABs-K199A, however, displays a loss-of-function in vivo (Fig. 7C), and the purified equivalent ComEAGs-K201A (Fig. S4) displayed only about a 2-fold decrease in apparent DNA binding affinity in vitro (Fig. 7E). We suspect that ComEAGs residue K201 (ComEABs residue K199) may mediate interactions between ComEA molecules when they bind DNA or participate in DNA binding interactions not apparent in the ComEAGs-DNA model.
While additional structural studies will be required to flesh out the details of the ComEA-ComEA and ComEA-DNA intermolecular interactions within a ComEA-DNA complex, ComEABs K164 appears to play a central role. Among all residues evaluated in our studies, ComEABs K164 is the most well-conserved throughout both the Gram-positives and Gram-negatives (Fig. 7A and S2). In fact, it is at least as well conserved in ComEA as the HhH motif signature residues. The equivalent V. cholerae residue, K63, was shown by the Blokesch lab to play an important role in that organism, and in silico modeling suggested it may contact DNA6, as it appears to do in our study (Fig. 7B).
In addition to pinpointing important residues for DNA binding, the EMSAs with wild-type ComEAGs and with the K201A mutant protein (Fig. 7E) exhibit evidence of cooperativity, consistent with OD-OD interactions between ComEA molecules bound to DNA. At intermediate concentrations of protein molecules, fully shifted DNA co-exists with unshifted and partially-shifted probe. A similar bimodal distribution in EMSA experiments has been reported for SSB, another non-specific DNA binding protein with documented cooperative binding behavior23.
Discussion
This study presents several notable findings. First, we have demonstrated the existence of a multimerization domain in ComEA from B. subtilis and G. stearothermophilus, also apparent in other multi-domain ComEA proteins encoded by the Gram-positives (Figs. S2 and 8A). Our AUC results confirm that the oligomerization interface deduced from the crystal structure is important for ComEA dimerization and the existence of ring structures in crystals of both ComEABs and ComEAGs, as well as an SV-AUC experiment with the isolated OD, show that ComEA may form extended multimers in vivo (Fig. 3A–D, Fig. 4B). As determined by MW-AUC, wild-type and ComEA-A108Y form DNA complexes with different molar ratios because DNA binding favors ComEA oligomerization by bringing the protein molecules into close approximation, thus providing a cooperative effect. The EMSA results displayed in Fig. 6E provide further strong support for cooperative DNA binding. While it is tempting to speculate that ComEA multimerization in vivo generates rings like those observed in the crystal structures, there is no evidence for this.
Finally, we have shown that both oligomerization and the DNA binding HhH motifs are needed for DNA uptake to the periplasm (Figs. 6 and 7). We have shown previously that ComEA plays important roles in B. subtilis4,18. Although the initial binding of tDNA to the transformable cell takes place to a surface exposed tpilus, this attachment is labile. When a loop of tDNA is pulled into the periplasm, presumably by disassembly of the tpilus filament9, the association of tDNA with the cell becomes stable as ComEA mediates DNA uptake to the periplasm18. In Neisseria gonorrheae and V. cholerae, the equivalent role of ComEA in uptake has been convincingly ascribed to a Brownian ratchet mechanism5,6,11,12,24, in which binding to DNA prevents retrograde diffusion. Now we have shown that although the DNA binding activity of ComEA is indeed needed for transformation, consistent with a simple ratchet, so is oligomerization, hinting at a more complex process than just the rectification of diffusion. An attractive and testable possibility is that simultaneous interaction of ComEA molecules with one another and with tDNA causes condensation of the DNA-protein complexes, providing a driving force for uptake (Fig. 8B). This would resemble the condensations recently reported for the interactions of DNA with the FoxA1 transcription factor and with the FUS protein25,26, both of which bind DNA as well as exhibiting self-interaction.
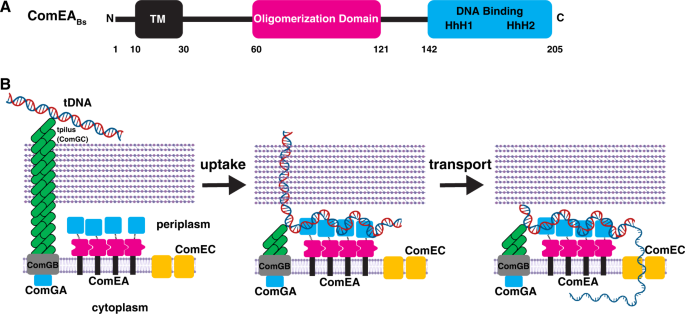
A ComEABs contains an N-terminal transmembrane region, a previously unidentified OD, and a C-terminal DNA-binding domain. B We have shown that ComEA self-associates using contacts in its ODs. After retraction of the tpilus to bring a loop of DNA into the periplasm, binding of tDNA to the ComEA DNA-binding domains followed by uptake stabilize cell association, while cross-linking of distal DNA segments by binding to adjacent ComEA molecules condenses the incoming tDNA, exerting a pulling force to bring tDNA into the periplasm. Elements of the figure were created with BioRender.com.
Consideration of ComEA-tDNA condensation as a force-generating machine must consider the geometry of ComEA within the periplasm. The DNA binding and oligomerization domains are connected by flexible linkers of 20–25 residues and the OD is separated from the periplasmic membrane surface by another linker of about 60 residues (Fig. 8A). Thus, there are considerable degrees of freedom for the two domains within the periplasm. This mobility may be an important component of the proposed condensation mechanism, permitting adjacent ComEA molecules to contact one another through their ODs while contacting different segments of tDNA through their DNA-binding domains, thus effectively cross-linking and condensing the tDNA (Fig. 8B). We propose therefore, that ComEA in Gram-positive bacteria may not be simply a Brownian ratchet but may function as a force-generating protein for the uptake of tDNA.
A comparison of the Gram-positive and Gram-negative ComEA molecules begs two questions: why has ComEABs evolved as a membrane protein while its single-domain orthologs are free to diffuse in the periplasm, and why do only the Gram-positive ComEA proteins contain an OD? The first question may be answered by the absence of an outer membrane in Gram-positives. Membrane anchoring will prevent the diffusion and loss of small proteins through the cell wall. Indeed, some ligand-binding proteins, e.g., those that bind amino acids for uptake, are diffusible in the periplasm of Gram-negative bacteria, but are membrane-anchored in the Gram-positives27,28,29. The presence of an OD only in the Gram-positives may also reflect a difference in surface structure compared to the Gram-negatives. In Gram-negative bacteria, diffusion of the transforming DNA through the characteristically thin cell wall may be rapid enough for a Brownian ratchet to effect efficient entry to the periplasm. But contacts of transforming DNA with the necessary channel in the thick peptidoglycan of Gram-positives, may provide a frictional impediment to diffusion, requiring additional pulling force. This intuitive concept is supported by a study showing that the rate of diffusion through a channel is reduced as the channel is made longer30.
Methods
ComEABs production
comEA (codons 58–205) was PCR amplified from genomic DNA from B. subtilis strain 168 using Phusion High-Fidelity DNA Polymerase (New England Biolabs) and His6-BsuF and His6-BsuR primers (Table S2). The amplified insert was cloned into the NcoI and XhoI restriction sites of pET28b with a His6 tag at its C-terminal end, generating expression vector pComEABs. E. coli BL21(D3E) transformed with pComEABs was grown at 37 °C in LB media to an OD of 0.6 in the presence of 30 μg/ml kanamycin with constant shaking at 200 rpm. ComEABs expression was then induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and grown overnight at 18 °C. The cells were then pelleted and resuspended in lysis Buffer A (30 mM Tris HCl [pH 8.0], 150 mM NaCl, 10 % glycerol) supplemented with 20 μg/ml DNase. The resuspended cells were lysed using a French press, and the lysate was clarified by centrifugation at 61,000 x g for 1 h at 4 °C. The clarified lysate was loaded on His-60 resin (Takara) equilibrated with Buffer B (30 mM Tris HCl [pH 8.0], 150 mM NaCl, 20 mM imidazole). The resin was washed with Buffer C (30 mM Tris HCl [pH 8.0], 300 mM NaCl, 20 mM imidazole) and then with Buffer B. To remove residual DNA, the resin was incubated with Buffer D (30 mM Tris HCl [pH 8.0], 150 mM NaCl, 10 mM imidazole, 1 mM MgCl2, 1 mM CaCl2 and 20 μg/ml DNase) at room temperature for 30 min. The resin was washed with Buffer B and the protein was eluted with Buffer B containing 500 mM imidazole. Protein purity was analyzed using SDS-PAGE. Fractions containing ComEABs were dialyzed overnight against Buffer E (30 mM Tris HCl [pH 8.0], 50 mM NaCl) and loaded onto a Source 15Q column equilibrated with Buffer E. The protein was eluted using a linear gradient of Buffer E and Buffer F (30 mM Tris HCl [pH 8.0], 1 M NaCl) over 20 column volumes. The fractions containing purified ComEABs were pooled and concentrated using a 3 kDa MWCO centrifugal concentrator device. The concentrated protein was loaded to a Superdex200 10/300 gel filtration column (GE Healthcare), which was equilibrated with Buffer G (15 mM citrate [pH 4.5], 100 mM NaCl). The eluted...
Comments
Post a Comment